Step 4. Identifying proteins
 When we searched for peptide identifications for our features, it's likely that some of our features were
matched to more than one peptide. Other features may have been assigned to the same peptide from
different proteins. These ambiguous peptide assignments are referred to as ‘conflicts’.
When we searched for peptide identifications for our features, it's likely that some of our features were
matched to more than one peptide. Other features may have been assigned to the same peptide from
different proteins. These ambiguous peptide assignments are referred to as ‘conflicts’.
To confidently identify our proteins, we need to resolve the ambiguity in our peptides' origins. The Protein View gives us the information we need to explore and resolve the conflicts. While Progenesis takes some steps to resolve conflicts automatically, you're likely to still be left with others than need to be resolved manually.
Resolving a simple conflict
Before we start, we'll focus the screen on the proteins with the most conflicts:
- Sort the Proteins list (top-left) on its Conflicts column by clicking the column header twice.
- Click the Protein Resolution tab in the lower half of the screen.
This will put the proteins with the most conflicting peptides at the top of the Proteins list (see the image above) and will allow us to see both sides of the selected conflict.
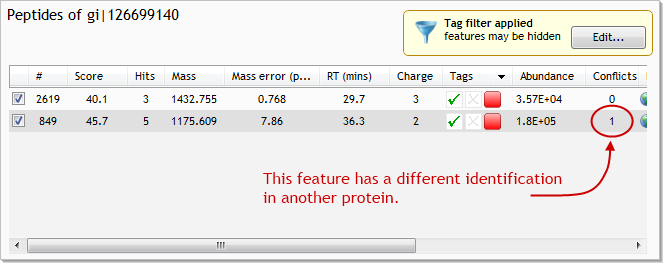
Now, in the Proteins list, find and select the protein with accession number gi|126699140. This protein has a single conflicting feature. Select the feature that has the conflict, being careful not to untick it at this stage (click the image below to see the full screen):
The list at the lower left of the screen now shows all identified proteins that contain the conflicting feature. In this case, there are only 2 such proteins: the one we selected (gi|126699140); and gi|126698718. By looking at the two proteins' lists of peptides, we can see that this conflict is, in fact, the only conflict that each protein has.
We now have to decide which peptide identification is more likely to be the true identification for the feature. As well as basing our decision on some of the numbers presented to us, we often also need to use our knowledge of the proteins themselves and their functions.
In this case, however, it's a relatively simple decision:
- there's greater evidence for the presence of gi|126698718, as it has matched 5 features, compared to the 2 features matched by gi|126699140
- the score returned from Mascot for the peptide in gi|126698718 is signficantly higher than that returned for the peptide in gi|126699140 (67.5 vs. 45.7)
- the mass error value for the peptide in gi|126698718 is significantly lower than that in gi|126699140
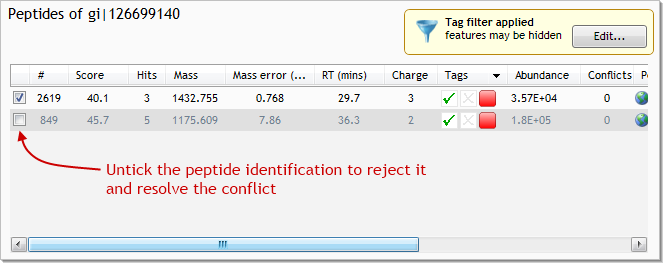
With these 3 pieces of evidence to back us up, we can reject the peptide identification from gi|126699140. To do so, untick the identification for feature #849 in gi|126699140:
To complete the analysis, we would continue resolving peptide conflicts like this, one protein at a time. As well as looking at search engine scores and the number of identified peptides, we can look at other properties such as individual peptide sequence lengths to help us decide which way a conflict should be resolved. We may even reject peptides from both proteins if the data is not decisive enough.
However, such a thorough analysis is beyond the scope of this tutorial. Instead, the following page summarises the analysis process and offers links to more in-depth guides for Progenesis LC‑MS…