Why do some of my peptide ions have identifications with no score?
This is because of the way Progenesis QI for proteomics carries out deconvolution of peptide ions for the combination of identification data across all the ions derived from a peptide.
Deconvolution allows the transfer of MS2 / MSE identification data across the various charge states of a peptide; this approach is used in order to provide the most complete information possible about a peptide’s identity. The result is that every peptide ion carries all the identification results from all other ions derived from the same peptide.
Where an identification result for one charge state is not present in another, it is transferred to the other as a result with a 'null' score (no score is entered). Therefore, when a peptide ion is shown with 'no score' for a given identification, you know that this particular identification has been derived from this process of deconvolution and identificaton-sharing.
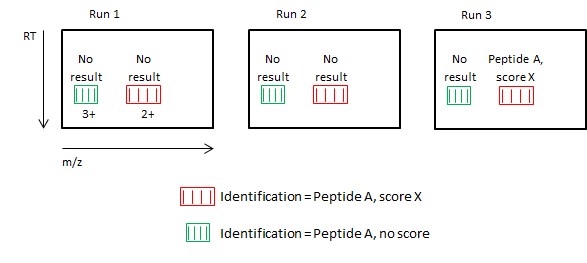
In the simplified illusration below:
- Each of the three runs has two peptide ions, 2+ and 3+.
- All ions are from the same peptide.
- Only the 2+ ion in Run 3 has an identification ("Peptide A" with score "X").
- First, the 2+ identification is transferred to the other 2+ ions in all runs (identification data for the same peptide ion are always combined as part of the no missing values approach).
- However, because deconvolution revealed that the two charge states are from the same peptide, the identification can be applied to 3+ ions as well.
- All ions have the identification "Peptide A", but the 3+ ions have a null score, indicating that the identification resulted from a deconvolution-based transfer.