Installing support for PLGS versions 2.4
Installing support for PLGS versions 2.4
To use this format you must install plugins to both Progenesis QI for proteomics and PLGS.
Adding the PLGS add-on to Progenesis QI for proteomics.
- Shut down Progenesis QI for proteomics.
- Download the Progenesis QI for proteomics add-on and double click the .msi file to install it.
- Open Progenesis QI for proteomics. In the “Identify Peptides” section, PLGS (*.xml) should now appear in the list of drop down options.
Adding the Progenesis plugin to your PLGS version 2.4 setup.
- Download the PLGS plugin file “nldplugin.jar”.
- Save it to the PLGS plugin folder, e.g. C:\PLGS2.4\jars\nldplugin.jar.
- Open the PLGS browser.
- Click the Options | Automation Setup menu item.
- Select the PlugIns tab.
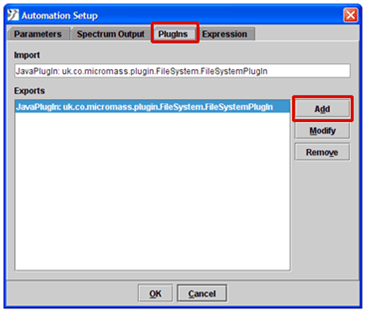
- Click the Add button.
- Select the Java Class option and enter the PlugIn Name and Class Path as shown below
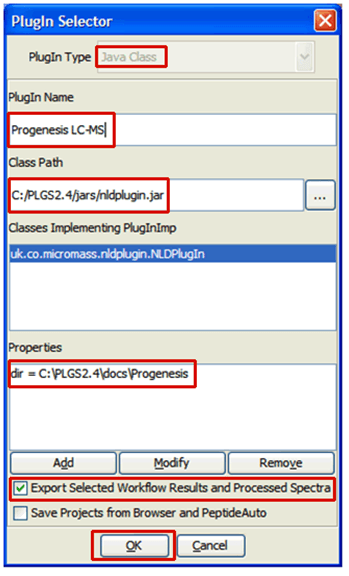
- Click the Add button bring up the Add/Modify properties dialog:
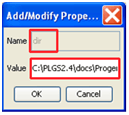
- Enter “dir” in the Name section
- Enter the folder you wish XML files to be exported to in the Value section:
- Click OK on the Add/Modify Properties, PlugIn Selector and Automation Setup dialogs.
- Close and restart the PLGS browser to register the changes.
See also
More peptide search formats supported by Progenesis QI for proteomics.





