How are the scores calculated for possible compound identifications?
Progenesis QI labels each identification with a "Score" to quantify the quality of the identification. These scores can be seen in the "Possible identifications" section of the "Identify Compounds" and "Review Compounds" screens:
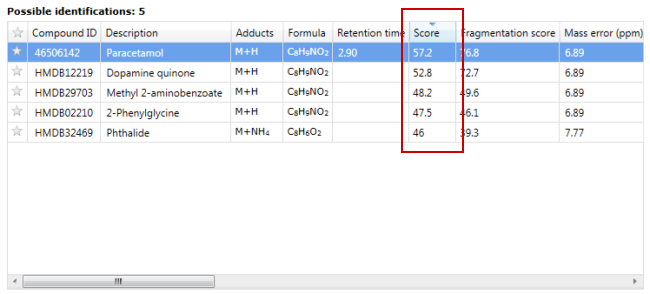
Scores range from 0-100, with 100 being a perfect match, and are calculated using the mean of five similarity metrics:
- Mass Similarity
- A mass difference of 0 gives a score of 100, which falls off rapidly to ~2 when the mass difference is ~1Da (for a 250Da compound).
- Isotope Similarity
- This compares the intensities of each isotope between observed and theoretical distributions. A total intensity difference of 0 gives a score of 100, which falls linearly to 0 when the total intensity difference is equal to the maximum isotope intensity.
- Retention Time Similarity
- A retention time difference of 0 gives a score of 100, which falls off rapidly to ~2 when the percentage error is ~20%. If the search method does not support searching by retention time or has been configured not to use it, this metric is 0.
- CCS Similarity
- A CCS difference of 0 gives a score of 100, which falls off rapidly to ~2 when the percentage error is ~20%. If the search method does not support searching by CCS or has been configured not to use it, this metric is 0.
- Fragmentation Score
- This is a score from 0 to 100 representing how well your observed fragmentation data matches the search (which might be either theoretical fragmentation, or data from a fragment database). The exact scoring method depends on whether you carried out a theoretical or fragment database search. If fragment search is not available or has been turned off in the configuration of your chosen search method, this metric is 0.
How is the fragmentation score calculated for possible compound identifications?
If you have chosen to use theoretical fragmentation, the fragmentation scores calculated for your compound are described in the FAQ on the theoretical fragmentation algorithm.
If you have chosen to compare your samples' fragmentation to the spectra held in a fragmentation database, the fragmentation scores calculated for your compound are described in the FAQ on the database fragmentation algorithm.






