Support for Bruker Daltonics .d folders

About this plug-in
You may also need to install the latest version of CompassXtract. This can be downloaded after logging in on the Bruker Daltonics' website.
This is the standard format produced by Bruker Daltonics mass spectrometers. Each sample run is contained within a separate .d folder, which typically contains a file named either analysis.baf or analysis.yep.
To select samples to import, first click the Browse button:
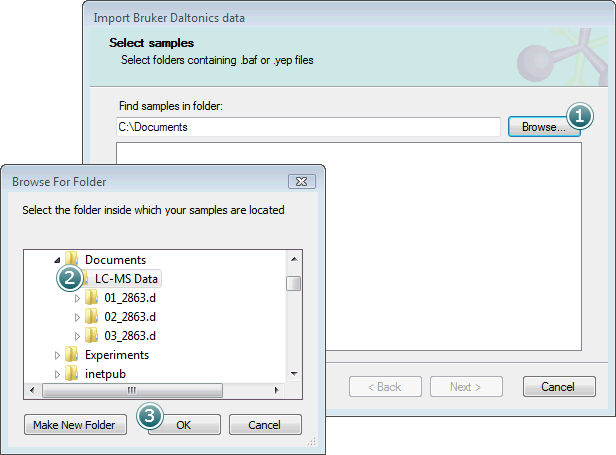
Select the parent folder containing your .d folders and click OK.
The .d folders within the parent will then be shown in the Select samples list box:
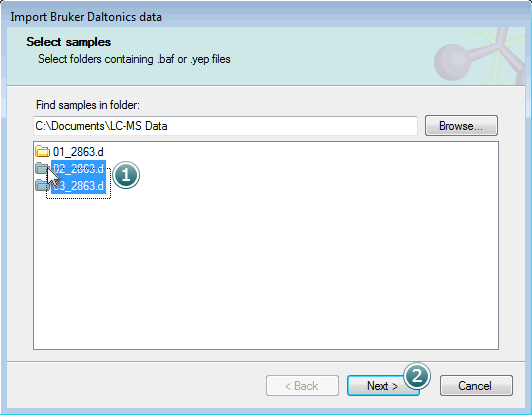
Select the samples you want to import and click Next to move on to the next step.
The next page allows you to include a linear m/z calibration as part of the import process:
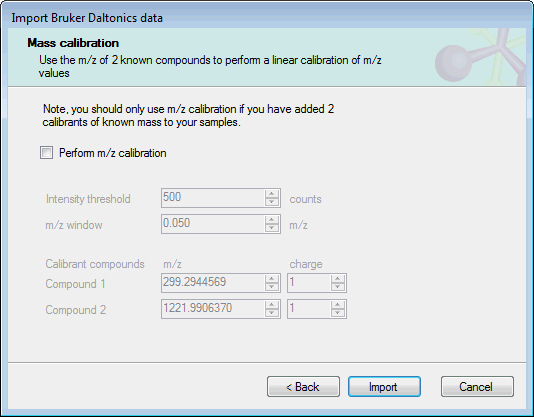
If you have included 2 calibrants of a known mass in your experimental samples, you can use these ions to calibrate the sample m/z values as they are imported. To do this, select Perform m/z calibration and set the expected m/z and charge for the calibrant ions. The other required option are:
- Intensity threshold: the minimum intensity in counts for a calibrant peak to be used in calibration.
- m/z window: the minimum m/z range over which the calibration algorithm should search for calibrant peaks.
If you do not want to apply an m/z calibration, clear the Perform m/z calibration checkbox.
Click the Import button to begin the data import.
This importer uses ProteoWizard to access the data in the Bruker Daltonics .d folders, and installs the ProteoWizard libaries as part of this data importer.





