How do I use tags?
 Tags are one of the most flexible analysis tools in Progenesis, allowing you to attach a label
to a selection of peptide ions. You can then show or hide peptide ions using a filter
that's based on a selection of their tags.
Tags are one of the most flexible analysis tools in Progenesis, allowing you to attach a label
to a selection of peptide ions. You can then show or hide peptide ions using a filter
that's based on a selection of their tags.
In this article:
- An example
- Tagging proteins
- Creating new tags
- Applying and removing tags
- Renaming and deleting tags
- Filtering your peptide ions
An example
For example, you could very quickly find peptide ions whose expression is increasing significantly by doing the following:
- Create a tag called "Significant" for all peptide ions that have a low p-value
- Create a tag called "High fold change" for all peptide ions that have a Fold value greater than 2
- Create a tag called "Up-regulated" for all peptide ions that have their highest values in a Treated condition
- Combine these tags in a filter, so that only peptide ions that have all three of the tags are shown
Tagging proteins
Although this article describes tagging the peptide ions in your experiment, the same approach can be used to tag
the identified proteins in your experiment. To avoid confusion Progenesis QI for proteomics displays protein tags in a circle  ,
and peptide ion tags are displayed in a square
,
and peptide ion tags are displayed in a square  .
.
Creating new tags
There are two ways to create new tags for your peptide ions:
- Create tags for the selected peptide ions
- Create tags for peptide ions having particular values
Creating a tag for the selected peptide ions
To create a tag for the selected peptide ions, first select the peptide ions to which you want to apply the tag. Features can be selected in the list at the left of the screen.
To select a range of peptide ions, you can click the peptide ion at the start of the range, then hold down the Shift key and click the peptide ion at the end of the range. To select or deselect an individual peptide ion, hold down the Ctrl key and click on it in the list.
Next, right-click on any of the selected peptide ions in the list. The tag menu will appear; from this, select the New Tag… option. The following window appears:
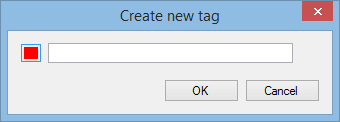
Enter the name that you want to give the tag. If you'd like it to have a different colour, click the coloured button to the left of the name. When happy, click OK. The tag is created and each of the selected peptide ions is given that tag. If any peptide ions already had a tag, the new tag is applied in addition to the existing tags.
Creating tags for peptide ions having particular values
Using the QuickTags feature, certain types of tag can be created without first having to select peptide ions. For example, you can quickly tag all peptide ions with a p-value of less than 0.05.
To use QuickTags, right-click anywhere in the list of peptide ions. In the tag menu, open the Quick Tags sub-menu and select one of the options in it. The New Quick Tag window will appear:
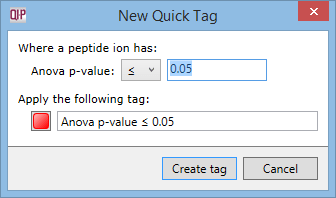
You can edit the name for the new tag, choose a different colour, or edit the criterion that will be used for applying the new tag. Then click Create tag. This time, the new tag will be applied to all peptide ions that meet the criterion entered in the New Quick Tag window.
Note: if your peptide ion measurements change, this tag will not be recalculated. That is, all peptide ions that were assigned the tag when it was first created will continue to have that tag, regardless of their new measurements.
Applying and removing tags
To give an existing tag to a peptide ion (or set of peptide ions), simply select the relevant peptide ions in the list, then either:
- right-click on any of the selected peptide ions, or
- click the arrow in the Tag column header
Either method will display the tag menu. From that, simply select the tag you wish to apply or remove.
Renaming and deleting tags
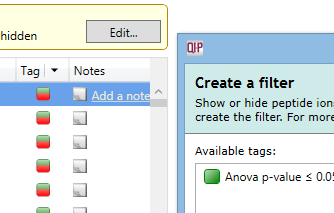
If you need to rename a tag, or you want to delete a tag that you no longer need in your experiment, this can be done easily. Click on the arrow in the Tag column header and select Edit tags from the menu. The Edit Tags window appears:
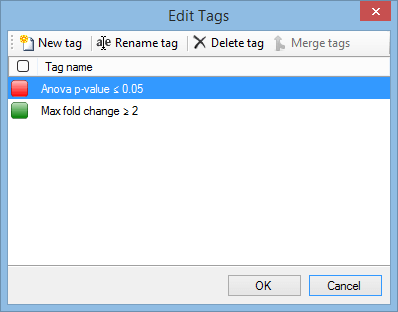
To rename a tag, highlight it and click Rename tag to show the Rename Tag window.
To delete a tag, highlight it and click Delete tag. Note that this will remove the tag from all peptide ions, not just the selected peptide ions.
Filtering your peptide ions
While labelling your peptide ions with tags is a helpful way to organize your data, filtering based on those tags is the more powerful analysis tool. By creating a filter, you can quickly reduce the amount of data you are viewing, enabling you to concentrate on the peptide ions of real interest.
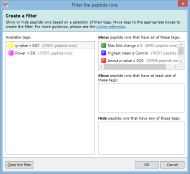
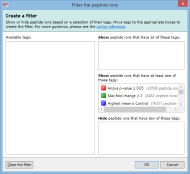
To create a filter, click the Create button in the filter panel shown above each peptide ion list. The Create a filter window appears:
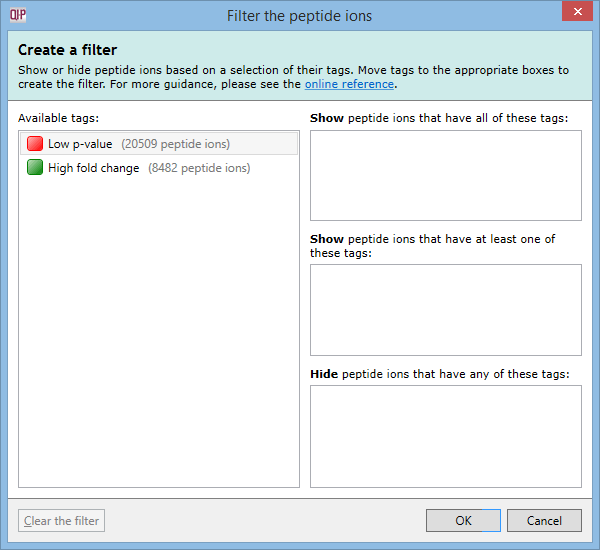
In this window, you can build a filter showing:
- peptide ions that have all of the tags in a given set
- peptide ions that have at least one of the tags in a given set
- peptide ions that have none of the tags in a given set
- or any combination of the above options
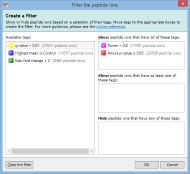
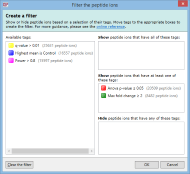
For example, imagine you have defined two tags: one for peptide ions that have a p-value of less than 0.05; and one for peptide ions that have a fold change of 2 or more. The Create a filter window would initially look as it does above. To show only the peptide ions that have both of the tags, click and drag each tag in the Available tags list to the top list on the right. When the done, the dialog looks like this:
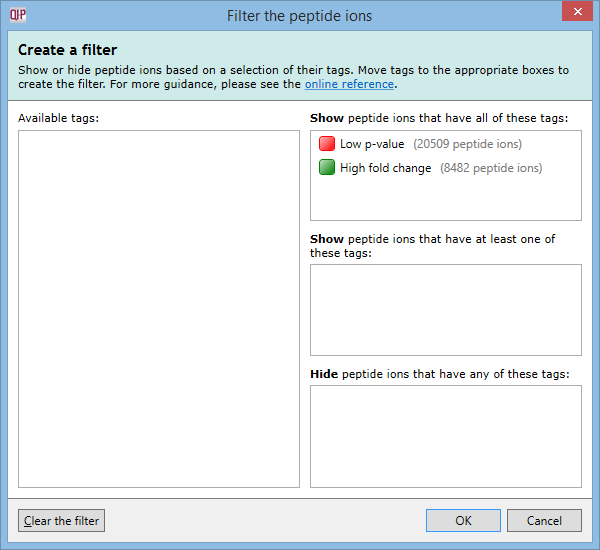
Only peptide ions that have both of these tags will be shown when you click OK. All other peptide ions will be hidden. Any tags left in the Available tags list will not affect the filter.
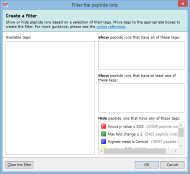
Here are some further examples of tag combinations you could use to filter your peptide ions:
Finally, to return to showing all peptide ions, just click the Clear the filter button and click OK.