How do I create my own spectral library?
You can create a spectral library from your own experiments using the following steps:
- Perform an analysis and search for identifications using one of the peptide search engines. (Note: if you are performing a search of Waters MSᴱ, HDMSᴱ or SONAR data using a search engine other than Ion Accounting, you must provide the exported MGF file when importing your results).
- Use the Refine Identifications and Resolve conflicts screens to filter identifications to only those you are confident are correct. This is an important step as Progenesis will export all possible identifications that haven't been deleted or rejected.
- If desired, apply a peptide ion tag filter to show only the ions whose identifications you wish to export to the spectral library.
-
At the Identify Peptides screen, choose Export spectral library… from the File menu.
- For DDA data, choose whether you wish to filter fragment ions in the dialog that is shown.
- Choose a file to export your library to.
- If exporting to an existing spectral library, you can choose to Overwrite or Append to the existing library.
How do I filter which fragment ions are exported to my spectral library?
Progenesis gives you the option to export full ms/ms spectra unfiltered, or filter the fragments based on the assigned sequences, so the spectral library will only contain relevant fragments.
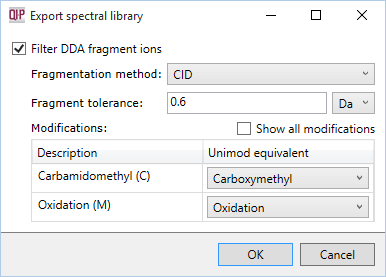
If you are exporting a spectral library from DDA data, you will see the following dialog:
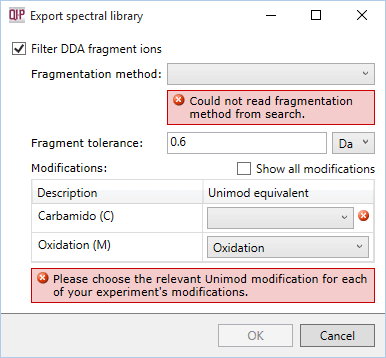
Progenesis will attempt to read the parameters from your search results. If this information is not available, you will see one or more error messages:
Progenesis filters fragments via the following method:
- For each identification, generate theoretical expected fragments based on the assigned sequence and any modifications.
- Compare the theoretical fragments to the experimental ms/ms spectrum. Only experimental fragments that match within the fragment tolerance are included in the spectral library.
Because this filtering process is performed in Progenesis, the resulting fragments may differ slightly to those shown by your search engine, e.g. Mascot.
Filtering parameters
- Filter DDA fragment ions
- If selected, fragment ion filtering will be performed. If not, the entire ms/ms spectrum will be exported to the spectral library for each identification.
- Fragmentation method
- Determines which ion types Progenesis considers when filtering fragment ions, and how PTMs behave during fragmentation. If the ECD fragmentation method is chosen, for example, Progenesis will ignore b ions, and will consider PTMs to always adhere during fragmentation.
- Fragment tolerance
- The m/z tolerance used when comparing experimental and theoretical fragments.
- Modifications
- Each modification found in your search results must be mapped to a known modification from UniMod. UniMod contains information about the modification such as its mass shift, specificity, and any possible neutral losses on fragmentation. For common modifications, Progenesis will attempt to perform this mapping automatically, but it may also require manual input. If you can't find the modification you are looking for in the list, select the Show all modifications check box.





