New features in Version 2.0 of Progenesis QI for proteomics
Progenesis QI for proteomics software enables you to Quantify and Identify proteins in your complex samples using the advantages of label-free analysis. With support for all common vendor data formats and a highly visual guided workflow, Progenesis QI for proteomics enables you to rapidly, objectively and reliably discover proteins of interest from single or fractionated samples, and using multi-group experimental designs. As well as conventional data-dependent analysis (DDA), Progenesis QI for proteomics supports the analysis of Waters MSE and HDMSE data-independent (DIA) data. Uniquely, the software also takes advantage of the additional dimension of resolution offered by Ion Mobility separations to give improvements in accuracy and precision of identification and quantification.
New Features
Pathway Analysis
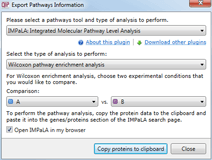
How do we understand the protein differences we have found in our experiment? One option is Pathway Analysis, which determines which biological pathways are implicated in the data, and thus provides the next level of information for biological contextualization. We have provided export tools to easily and quickly interface with third party Pathway Analysis programs.
Easy export functionality to Pathway Analysis tools facilitates the process of placing discoveries into a biological context and extracting maximum value from your OMICS data.
QC Metrics
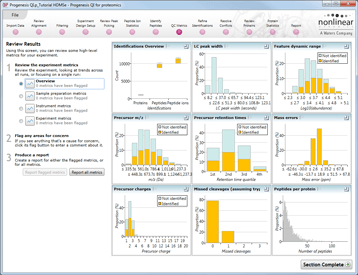
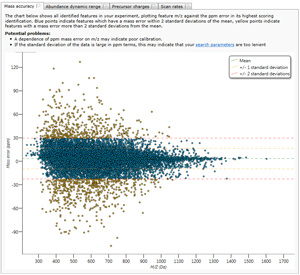
QC Metric tools aim to avoid wasting your valuable time performing data analysis on sub-optimal LC-MS data and include readouts such as LC peak width, feature dynamic range, precursor mass error, missed cleavage count and peptide per protein count. In addition, these QC Metric tools can be used to guide process optimization and troubleshooting.
QC Metrics are available for a range of processes from sample preparation to instrument operation.
Automation of Process Steps
To free up your time for other important tasks and maximize the opportunity for overnight processing, we have introduced additional functionality to enable the automation of many analysis steps. It is now possible to move from Data Import to a completed Protein Database Search without any external user intervention.
Improved 'HiN' Quantification
Quantification is an important element of any proteomics study. We have enhanced the 'Hi3' absolute quantification method in previous versions by allowing the user to define the number of peptides to be used for protein quantification. This functionality is also extended to 2D-LC experiments.
Speed and Usability
To improve usability, the Review Peak Picking window has received a full rebuild to increase the responsiveness to user queries and edits.