Support for METLIN™ MS/MS Library 2019
For details of pricing and how to purchase this compound search method, please contact us.
About this plug-in
The Waters® METLIN™ MS/MS Library 2019 for Progenesis QI contains a local copy of the METLIN database, and allows you to search this copy rapidly.
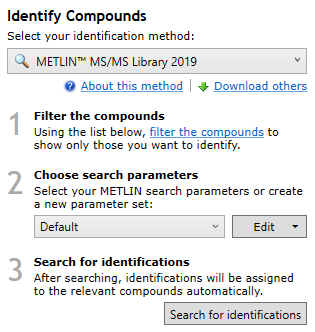
The METLIN™ MS/MS Library 2019 search method, with example parameters chosen.
The use of this method has the following steps:
- In the Identify Compounds screen, filter the list of compounds to show only those that you want to identify.
- Choose a set of search parameters from the Choose search parameters drop-down (you might need to first create or edit a search parameter set. See the section on search parameters below for how to do this).
- Click the Search for identifications button.
When the search is complete, a prompt will tell you how many compounds have been identified. You can then continue with your analysis, identifying further compounds if necessary by searching with other parameter sets.
Search parameters
The Waters® METLIN™ MS/MS Library 2019 for Progenesis QI comes with a basic set of default parameters. It will search the library with a precursor tolerance of 12ppm and a fragment tolerance of 12ppm. However, you may wish to create your own parameter sets, or modify the built in configuration.
To edit or create a new search parameter set, choose the appropriate option in the drop-down to the right of the parameter set name:
This will open a dialog where you can fill in the various search parameters:
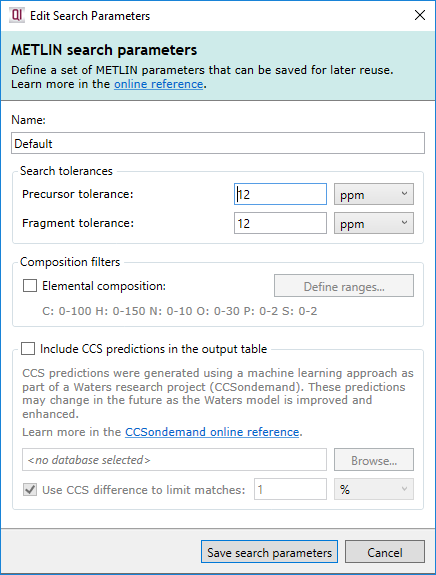
Briefly, the core options available to you are:
- Name
- Give the search parameter set a useful name to refer to later.
- Precursor tolerance
- Choose the threshold (in ppm or Da) within which the neutral mass must lie to constitute a match.
- Fragment tolerance
- Choose the threshold (in ppm or Da) within which fragments must match the database to contribute to the score.
It is also possible to further filter the identifications using elemental composition. By ticking the Filter by elemental composition checkbox and clicking the Select elemental composition... button, you will see the following dialog:
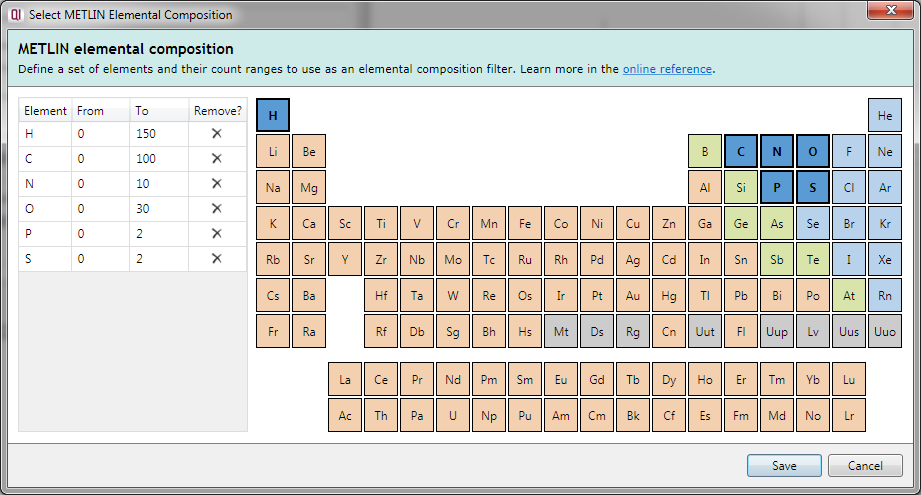
Clicking on a currently-excluded element in the periodic table table adds it to the filter, allowing compounds containing that element to be considered as possible identifications. The element will now be listed in the table on the left.
The From and To fields in the table allow you to specify a minimum and maximum number of atoms of each element that may be present in the compound.
To remove an element from the specification, you can press the Delete key with that element selected in the left table, click on the cross button in the Remove? column of that table, or click on the element in the periodic table a second time.
After entering your search parameters, click Save search parameters to save your search parameter set for use in this and future experiments.
Licensing
Before using the METLIN™ MS/MS Library 2019 for Progenesis QI, you need to activate a METLIN™ MS/MS Library 2019 for Progenesis QI license on your existing Progenesis dongle.
All subsequent uses of the METLIN™ MS/MS Library 2019 for Progenesis QI require the activated dongle to be plugged in.
You can find out how to activate your METLIN™ MS/MS Library 2019 for Progenesis QI license here.
CCSondemand
CCSondemand is an optional feature that is available on request. Contact us for more information if you are interested in this feature.
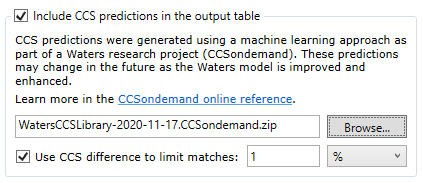
This feature uses CCS predictions which were generated using a machine learning approach as part of the Waters CCSondemand research project. These predictions may change in the future as the Waters model is improved and enhanced.
As the predictions are available for a limited set of adducts: +H, -H, +Na, +K, +HCOO, you will only be able to get CCS predictions for compounds that Progenesis has detected as having these adducts.
Prediction errors, are in most instances, of the same order of magnitude as experimental intra-instrument CCS measurement variation and can be used as a surrogate for cases without measurements from standards.
Upon request, we can provide you with a link to download a zip file containing the supplemental CCS data required by this feature. This zip file is encrypted and you do not need to unzip it. Tell Progenesis the location of this zip file by using the Browse... button to locate the file on your PC. You can then use the checkboxes to control whether you get CCS annotations of your search results, and whether the difference between predicted CCS and measured CCS will be used in scoring matches.
To include CCS annotations in your search results
Check the box Include CCS predictions in the output table to annotate your search results with CCS predictions.
To filter out search results that exceed a certain CCS tolerance
Check the box Use CCS difference to limit matches to exclude matches where CCS predictions are available and the difference between the observed CCS and the prediction exceeds the specified tolerance.