Support for MetaCore, a Cortellis solution
Support for this pathway analysis tool is provided as standard.
About this plug-in
MetaCore, a Cortellis solution, is one solution in the Cortellis™ suite of intelligence solutions that enables precise, actionable answers to specific questions across the R&D lifecycle. MetaCore delivers high-quality systems biology content in the context of comprehensive pathway-based analytics, molecular network building and insightful visualizations.
Visit the Cortellis™ website to learn more.
Using the tool
Firstly, you will need a MetaCore license to use his tool, and you will also need your MetaCore account login information to export data from Progenesis to MetaCore.
The export works with identifications matching the current tag filter.
To begin, select Export to pathways tool from the Review Compounds screen, then select MetaCore from the Which pathways tool do you want to use? drop-down list.
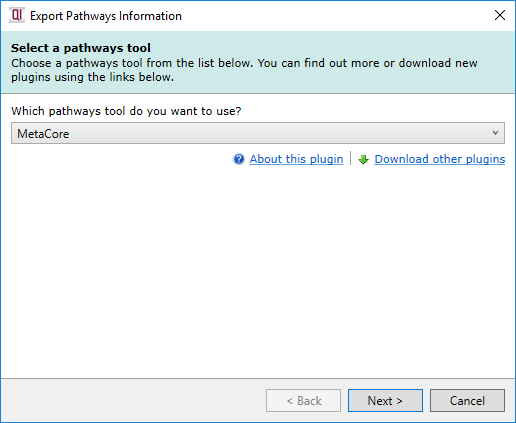
Selecting the MetaCore pathways tool.
Click Next > to move to the Configure your export page.
You can select the appropriate identifier type here.
If you have multiple conditions in your current experiment design, you can select which condition should be used as the control. Progenesis will calculate the abundance fold change and p-value for every other condition compared to the nominated control condition, and this data will be exported to MetaCore.
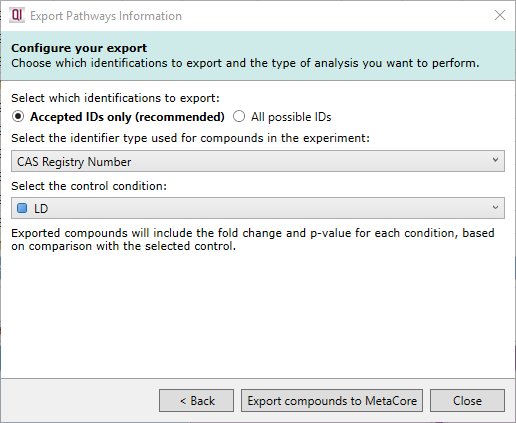
Selecting identifier type and control condition.
When you are happy with these settings click Export compounds to MetaCore.
If you have not already done so, you will be prompted to enter your MetaCore User name and Password; once you have done this, your data will be exported to your MetaCore account, and the MetaCore user interface will open in your default web-browser.
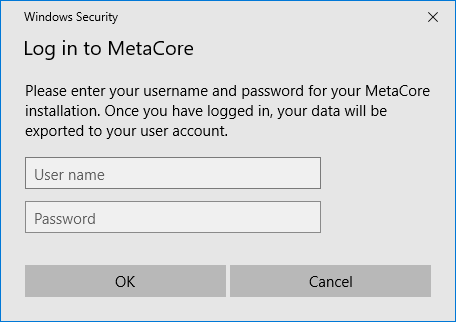
Providing your MetaCore account details.




