Support for Progenesis MetaScope

About this plug-in
This identification method is intended to support identifications from a number of different compound databases. It allows you to choose a .SDF, .CSV, .XLS or .XLSX file containing identifications and information on a selection of compounds. Features are then matched to m/z and/or retention times specified in these data files.
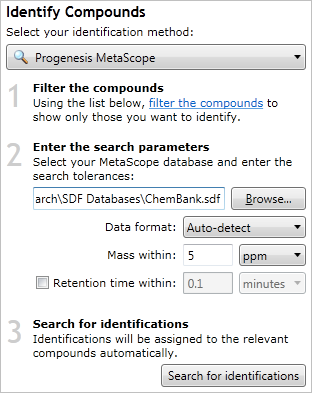
The MetaScope search method, with an example file and parameters chosen.
The use of this method has the following steps:
- In the Identify Compounds screen, filter the list of features to show only those that you want to identify.
- Click the Browse… button and choose the .SDF, .CSV, .XLS or .XLSX file you wish to use for searching.
- If you have chosen a .SDF file, and you know which database it has been exported from, choose the database name in the Data format dropdown. This gives MetaScope further information on how to read information from this specific .SDF file. If you don't know the origin of the .SDF file, just choose Auto-detect.
- Choose the mass range within which a positive identification will be made in the Mass within field, or leave the default of 5 ppm.
- If you wish to also match on retention time, tick the Retention time within check box, and choose a retention time window.
- Click the Search for identifications button.
When the import and search is complete, a prompt will tell you how many features have been identified. You can then continue with your analysis, identifying further compounds if necessary by searching other database files.
SDF Files
The SDF format contains structural information and other associated data about the metabolites in the database.
Here are some good sources of metabolite SDF databases which can be searched with MetaScope:
- HMDB (Note: you need version 1.0.4843.40922 or later of the MetaScope plugin to be able to read this file. Please contact support if you have any problems.)
- YMDB
- LipidMaps
- DrugBank
- CSF Metabolome
Excel File Format
If you are using a .CSV, .XLS or .XLSX file for MetaScope identifications, it should contain the following fields:
| Field Name | Required? |
|---|---|
| Neutral Mass | Yes |
| Compound Id | Yes |
| Description | No |
| Formula | No |
| Url | No |
| Retention Time (min) | If Retention time within checked. |
Field names are case insensitive and can occur in any order.
You can also download an example database.
Note: you will need to have Microsoft Excel 2007 (or later) installed to perform the search.
Identification Scores
MetaScope labels each identification with a "Score" to quantify the quality of the identification. These scores can be seen in the "Possible identifications" tab of the "Review Compounds" screen.
Scores range from 0-100, with 100 being a perfect match, and are calculated using the mean of three similarity metrics:
- Mass Similarity
- A mass difference of 0 gives a score of 100, which falls off rapidly to ~2 when the mass difference is ~1Da (for a 250Da compound).
- Retention Time Similarity
- A retention time difference of 0 gives a score of 100, which falls off rapidly to ~2 when the percentage error is ~20%. If the search didn't have Retention time within checked, this metric is 0.
- Isotope Similarity
- This compares the intensities of each isotope between observed and theoretical distributions. A total intentity difference of 0 gives a score of 100, which falls linearly to 0 when the total intensity difference is equal to the maximum isotope intensity.





