How can I export my compounds' isotope profiles?
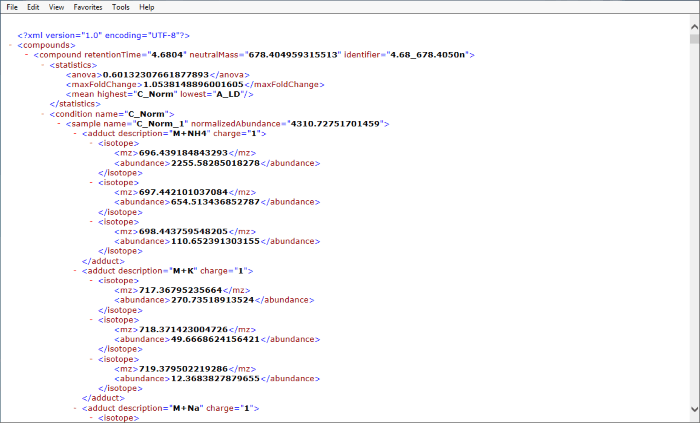
Upon selecting the File | Export isotope information… option at the Identify Compounds or Review Compounds stages, you will be given a pop-up dialog prompting you to name and save an xml file. The file generated will contain information for each compound, as follows:
Initial information – aggregate level
- Retention time
- Neutral mass
- Compound identifier
- ANOVA (for current experimental design)
- Maximum fold change (in normalised abundance, between any two groups in the current experiment design)
- Highest and lowest mean (of normalised abundance, of any conditions in current experiment design)
Isotope details – by sample, grouped in conditions
Within sample, compound level:
- Sample name
- Compound normalised abundance
Within compound, adduct level:
- Adduct description (the adducted species)
- Adduct charge
Within adduct, isotope level:
- Per-isotope m/z
- Per-isotope abundance
An example is shown below (click to expand):
Screenshot of an xml export resulting from the Export isotope information option as viewed in IE 11.